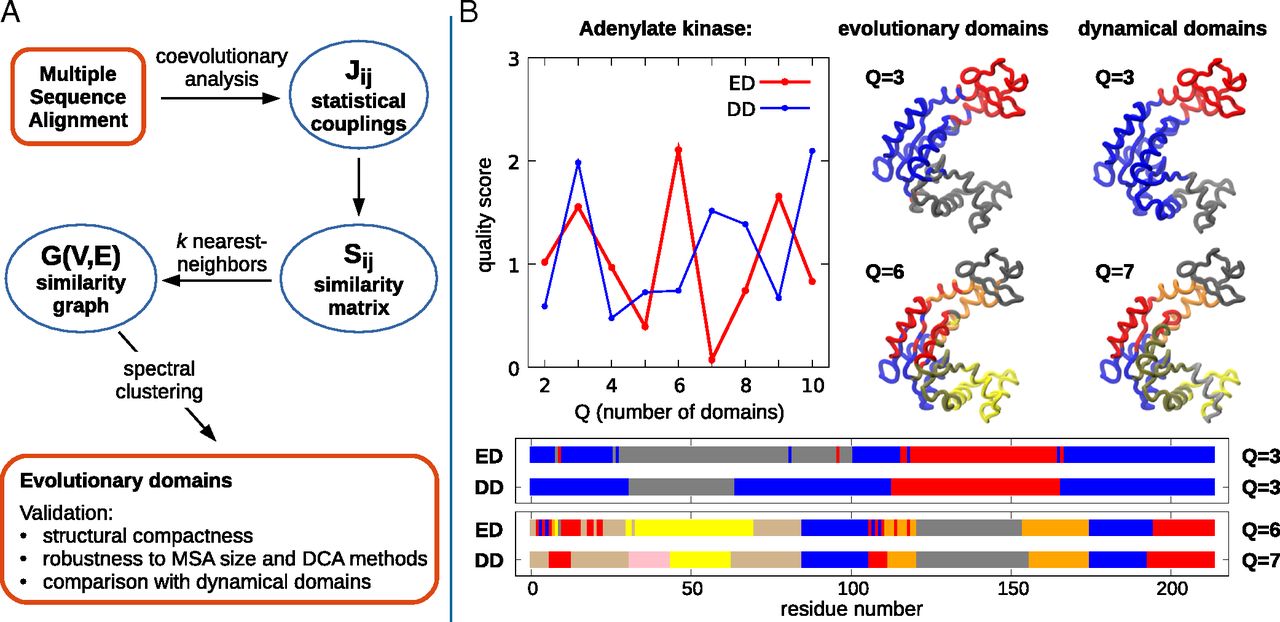
SPECTRUS Web Server
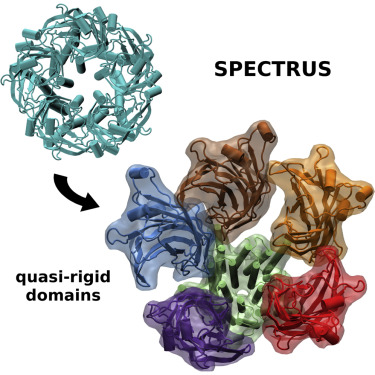
The SPECTRUS web server (Spectral-based Rigid Unit Subdivision) performs a decomposition into quasi-rigid
domains of proteins or protein complexes, based on the analysis of the distance fluctuations between pairs of
amino acids. The server processes entries of up to 2000 amino acids and trajectory files of up to 50MB. If your
data exceed this limit, please consider to run SPECTRUS locally on your machine by installing the source code,
available here.
Ponzoni L, Polles G, Carnevale V, Micheletti C, "SPECTRUS: a dimensionality reduction approach for identifying
dynamical domains in protein complexes from limited structural datasets", Structure, 2015.